models.tfidfmodel – TF-IDF model¶
This module implements functionality related to the Term Frequency - Inverse Document Frequency class of bag-of-words vector space models.
- class gensim.models.tfidfmodel.TfidfModel(corpus=None, id2word=None, dictionary=None, wlocal=<function identity>, wglobal=<function df2idf>, normalize=True, smartirs=None, pivot=None, slope=0.25)¶
Bases:
TransformationABCObjects of this class realize the transformation between word-document co-occurrence matrix (int) into a locally/globally weighted TF-IDF matrix (positive floats).
Examples
>>> import gensim.downloader as api >>> from gensim.models import TfidfModel >>> from gensim.corpora import Dictionary >>> >>> dataset = api.load("text8") >>> dct = Dictionary(dataset) # fit dictionary >>> corpus = [dct.doc2bow(line) for line in dataset] # convert corpus to BoW format >>> >>> model = TfidfModel(corpus) # fit model >>> vector = model[corpus[0]] # apply model to the first corpus document
Compute TF-IDF by multiplying a local component (term frequency) with a global component (inverse document frequency), and normalizing the resulting documents to unit length. Formula for non-normalized weight of term
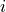 in document
in document 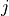 in a corpus of
in a corpus of 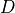 documents
documents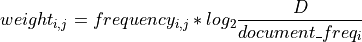
or, more generally
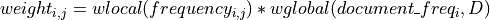
so you can plug in your own custom
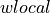 and
and 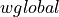 functions.
functions.- Parameters
corpus (iterable of iterable of (int, int), optional) – Input corpus
id2word ({dict,
Dictionary}, optional) – Mapping token - id, that was used for converting input data to bag of words format.dictionary (
Dictionary) – If dictionary is specified, it must be a corpora.Dictionary object and it will be used. to directly construct the inverse document frequency mapping (then corpus, if specified, is ignored).wlocals (callable, optional) – Function for local weighting, default for wlocal is
identity()(other options:numpy.sqrt(), lambda tf: 0.5 + (0.5 * tf / tf.max()), etc.).wglobal (callable, optional) – Function for global weighting, default is
df2idf().normalize ({bool, callable}, optional) – Normalize document vectors to unit euclidean length? You can also inject your own function into normalize.
smartirs (str, optional) –
SMART (System for the Mechanical Analysis and Retrieval of Text) Information Retrieval System, a mnemonic scheme for denoting tf-idf weighting variants in the vector space model. The mnemonic for representing a combination of weights takes the form XYZ, for example ‘ntc’, ‘bpn’ and so on, where the letters represents the term weighting of the document vector.
- Term frequency weighing:
b - binary,
t or n - raw,
a - augmented,
l - logarithm,
d - double logarithm,
L - log average.
- Document frequency weighting:
x or n - none,
f - idf,
t - zero-corrected idf,
p - probabilistic idf.
- Document normalization:
x or n - none,
c - cosine,
u - pivoted unique,
b - pivoted character length.
Default is ‘nfc’. For more information visit SMART Information Retrieval System.
pivot (float or None, optional) –
In information retrieval, TF-IDF is biased against long documents 1. Pivoted document length normalization solves this problem by changing the norm of a document to slope * old_norm + (1.0 - slope) * pivot.
You can either set the pivot by hand, or you can let Gensim figure it out automatically with the following two steps:
Set either the u or b document normalization in the smartirs parameter.
Set either the corpus or dictionary parameter. The pivot will be automatically determined from the properties of the corpus or dictionary.
If pivot is None and you don’t follow steps 1 and 2, then pivoted document length normalization will be disabled. Default is None.
See also the blog post at https://rare-technologies.com/pivoted-document-length-normalisation/.
slope (float, optional) –
In information retrieval, TF-IDF is biased against long documents 1. Pivoted document length normalization solves this problem by changing the norm of a document to slope * old_norm + (1.0 - slope) * pivot.
Setting the slope to 0.0 uses only the pivot as the norm, and setting the slope to 1.0 effectively disables pivoted document length normalization. Singhal 2 suggests setting the slope between 0.2 and 0.3 for best results. Default is 0.25.
See also the blog post at https://rare-technologies.com/pivoted-document-length-normalisation/.
References
- 1(1,2)
Singhal, A., Buckley, C., & Mitra, M. (1996). Pivoted Document Length Normalization. SIGIR Forum, 51, 176–184.
- 2
Singhal, A. (2001). Modern information retrieval: A brief overview. IEEE Data Eng. Bull., 24(4), 35–43.
- __getitem__(bow, eps=1e-12)¶
Get the tf-idf representation of an input vector and/or corpus.
- bow{list of (int, int), iterable of iterable of (int, int)}
Input document in the sparse Gensim bag-of-words format, or a streamed corpus of such documents.
- epsfloat
Threshold value, will remove all position that have tfidf-value less than eps.
- Returns
vector (list of (int, float)) – TfIdf vector, if bow is a single document
TransformedCorpus– TfIdf corpus, if bow is a corpus.
- add_lifecycle_event(event_name, log_level=20, **event)¶
Append an event into the lifecycle_events attribute of this object, and also optionally log the event at log_level.
Events are important moments during the object’s life, such as “model created”, “model saved”, “model loaded”, etc.
The lifecycle_events attribute is persisted across object’s
save()andload()operations. It has no impact on the use of the model, but is useful during debugging and support.Set self.lifecycle_events = None to disable this behaviour. Calls to add_lifecycle_event() will not record events into self.lifecycle_events then.
- Parameters
event_name (str) – Name of the event. Can be any label, e.g. “created”, “stored” etc.
event (dict) –
Key-value mapping to append to self.lifecycle_events. Should be JSON-serializable, so keep it simple. Can be empty.
This method will automatically add the following key-values to event, so you don’t have to specify them:
datetime: the current date & time
gensim: the current Gensim version
python: the current Python version
platform: the current platform
event: the name of this event
log_level (int) – Also log the complete event dict, at the specified log level. Set to False to not log at all.
- initialize(corpus)¶
Compute inverse document weights, which will be used to modify term frequencies for documents.
- Parameters
corpus (iterable of iterable of (int, int)) – Input corpus.
- classmethod load(*args, **kwargs)¶
Load a previously saved TfidfModel class. Handles backwards compatibility from older TfidfModel versions which did not use pivoted document normalization.
- save(fname_or_handle, separately=None, sep_limit=10485760, ignore=frozenset({}), pickle_protocol=4)¶
Save the object to a file.
- Parameters
fname_or_handle (str or file-like) – Path to output file or already opened file-like object. If the object is a file handle, no special array handling will be performed, all attributes will be saved to the same file.
separately (list of str or None, optional) –
If None, automatically detect large numpy/scipy.sparse arrays in the object being stored, and store them into separate files. This prevent memory errors for large objects, and also allows memory-mapping the large arrays for efficient loading and sharing the large arrays in RAM between multiple processes.
If list of str: store these attributes into separate files. The automated size check is not performed in this case.
sep_limit (int, optional) – Don’t store arrays smaller than this separately. In bytes.
ignore (frozenset of str, optional) – Attributes that shouldn’t be stored at all.
pickle_protocol (int, optional) – Protocol number for pickle.
See also
load()Load object from file.
- gensim.models.tfidfmodel.df2idf(docfreq, totaldocs, log_base=2.0, add=0.0)¶
Compute inverse-document-frequency for a term with the given document frequency docfreq:
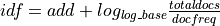
- Parameters
docfreq ({int, float}) – Document frequency.
totaldocs (int) – Total number of documents.
log_base (float, optional) – Base of logarithm.
add (float, optional) – Offset.
- Returns
Inverse document frequency.
- Return type
float
- gensim.models.tfidfmodel.precompute_idfs(wglobal, dfs, total_docs)¶
Pre-compute the inverse document frequency mapping for all terms.
- Parameters
wglobal (function) – Custom function for calculating the “global” weighting function. See for example the SMART alternatives under
smartirs_wglobal().dfs (dict) – Dictionary mapping term_id into how many documents did that term appear in.
total_docs (int) – Total number of documents.
- Returns
Inverse document frequencies in the format {term_id_1: idfs_1, term_id_2: idfs_2, …}.
- Return type
dict of (int, float)
- gensim.models.tfidfmodel.resolve_weights(smartirs)¶
Check the validity of smartirs parameters.
- Parameters
smartirs (str) –
smartirs or SMART (System for the Mechanical Analysis and Retrieval of Text) Information Retrieval System, a mnemonic scheme for denoting tf-idf weighting variants in the vector space model. The mnemonic for representing a combination of weights takes the form ddd, where the letters represents the term weighting of the document vector. for more information visit SMART Information Retrieval System.
- Returns
str of (local_letter, global_letter, normalization_letter)
local_letter (str) –
- Term frequency weighing, one of:
b - binary,
t or n - raw,
a - augmented,
l - logarithm,
d - double logarithm,
L - log average.
global_letter (str) –
- Document frequency weighting, one of:
x or n - none,
f - idf,
t - zero-corrected idf,
p - probabilistic idf.
normalization_letter (str) –
- Document normalization, one of:
x or n - none,
c - cosine,
u - pivoted unique,
b - pivoted character length.
- Raises
ValueError – If smartirs is not a string of length 3 or one of the decomposed value doesn’t fit the list of permissible values.
- gensim.models.tfidfmodel.smartirs_normalize(x, norm_scheme, return_norm=False)¶
Normalize a vector using the normalization scheme specified in norm_scheme.
- Parameters
x (numpy.ndarray) – The tf-idf vector.
norm_scheme ({'n', 'c'}) – Document length normalization scheme.
return_norm (bool, optional) – Return the length of x as well?
- Returns
numpy.ndarray – Normalized array.
float (only if return_norm is set) – Norm of x.
- gensim.models.tfidfmodel.smartirs_wglobal(docfreq, totaldocs, global_scheme)¶
Calculate global document weight based on the weighting scheme specified in global_scheme.
- Parameters
docfreq (int) – Document frequency.
totaldocs (int) – Total number of documents.
global_scheme ({'n', 'f', 't', 'p'}) – Global transformation scheme.
- Returns
Calculated global weight.
- Return type
float
- gensim.models.tfidfmodel.smartirs_wlocal(tf, local_scheme)¶
Calculate local term weight for a term using the weighting scheme specified in local_scheme.
- Parameters
tf (int) – Term frequency.
local ({'b', 'n', 'a', 'l', 'd', 'L'}) – Local transformation scheme.
- Returns
Calculated local weight.
- Return type
float
