corpora.indexedcorpus – Random access to corpus documents¶
Base Indexed Corpus class.
- class gensim.corpora.indexedcorpus.IndexedCorpus(fname, index_fname=None)¶
Bases:
CorpusABCIndexed corpus is a mechanism for random-accessing corpora.
While the standard corpus interface in gensim allows iterating over corpus, we’ll show it with
MmCorpus.>>> from gensim.corpora import MmCorpus >>> from gensim.test.utils import datapath >>> >>> corpus = MmCorpus(datapath('testcorpus.mm')) >>> for doc in corpus: ... pass
IndexedCorpusallows accessing the documents with index in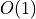 look-up time.
look-up time.>>> document_index = 3 >>> doc = corpus[document_index]
Notes
This functionality is achieved by storing an extra file (by default named the same as the fname.index) that stores the byte offset of the beginning of each document.
- Parameters
fname (str) – Path to corpus.
index_fname (str, optional) – Path to index, if not provided - used fname.index.
- add_lifecycle_event(event_name, log_level=20, **event)¶
Append an event into the lifecycle_events attribute of this object, and also optionally log the event at log_level.
Events are important moments during the object’s life, such as “model created”, “model saved”, “model loaded”, etc.
The lifecycle_events attribute is persisted across object’s
save()andload()operations. It has no impact on the use of the model, but is useful during debugging and support.Set self.lifecycle_events = None to disable this behaviour. Calls to add_lifecycle_event() will not record events into self.lifecycle_events then.
- Parameters
event_name (str) – Name of the event. Can be any label, e.g. “created”, “stored” etc.
event (dict) –
Key-value mapping to append to self.lifecycle_events. Should be JSON-serializable, so keep it simple. Can be empty.
This method will automatically add the following key-values to event, so you don’t have to specify them:
datetime: the current date & time
gensim: the current Gensim version
python: the current Python version
platform: the current platform
event: the name of this event
log_level (int) – Also log the complete event dict, at the specified log level. Set to False to not log at all.
- classmethod load(fname, mmap=None)¶
Load an object previously saved using
save()from a file.- Parameters
fname (str) – Path to file that contains needed object.
mmap (str, optional) – Memory-map option. If the object was saved with large arrays stored separately, you can load these arrays via mmap (shared memory) using mmap=’r’. If the file being loaded is compressed (either ‘.gz’ or ‘.bz2’), then `mmap=None must be set.
See also
save()Save object to file.
- Returns
Object loaded from fname.
- Return type
object
- Raises
AttributeError – When called on an object instance instead of class (this is a class method).
- save(*args, **kwargs)¶
Saves the in-memory state of the corpus (pickles the object).
Warning
This saves only the “internal state” of the corpus object, not the corpus data!
To save the corpus data, use the serialize method of your desired output format instead, e.g.
gensim.corpora.mmcorpus.MmCorpus.serialize().
- static save_corpus(fname, corpus, id2word=None, metadata=False)¶
Save corpus to disk.
Some formats support saving the dictionary (feature_id -> word mapping), which can be provided by the optional id2word parameter.
Notes
Some corpora also support random access via document indexing, so that the documents on disk can be accessed in O(1) time (see the
gensim.corpora.indexedcorpus.IndexedCorpusbase class).In this case,
save_corpus()is automatically called internally byserialize(), which doessave_corpus()plus saves the index at the same time.Calling
serialize() is preferred to calling :meth:`gensim.interfaces.CorpusABC.save_corpus().- Parameters
fname (str) – Path to output file.
corpus (iterable of list of (int, number)) – Corpus in BoW format.
id2word (
Dictionary, optional) – Dictionary of corpus.metadata (bool, optional) – Write additional metadata to a separate too?
- classmethod serialize(fname, corpus, id2word=None, index_fname=None, progress_cnt=None, labels=None, metadata=False)¶
Serialize corpus with offset metadata, allows to use direct indexes after loading.
- Parameters
fname (str) – Path to output file.
corpus (iterable of iterable of (int, float)) – Corpus in BoW format.
id2word (dict of (str, str), optional) – Mapping id -> word.
index_fname (str, optional) – Where to save resulting index, if None - store index to fname.index.
progress_cnt (int, optional) – Number of documents after which progress info is printed.
labels (bool, optional) – If True - ignore first column (class labels).
metadata (bool, optional) – If True - ensure that serialize will write out article titles to a pickle file.
Examples
>>> from gensim.corpora import MmCorpus >>> from gensim.test.utils import get_tmpfile >>> >>> corpus = [[(1, 0.3), (2, 0.1)], [(1, 0.1)], [(2, 0.3)]] >>> output_fname = get_tmpfile("test.mm") >>> >>> MmCorpus.serialize(output_fname, corpus) >>> mm = MmCorpus(output_fname) # `mm` document stream now has random access >>> print(mm[1]) # retrieve document no. 42, etc. [(1, 0.1)]
