Note
Click here to download the full example code
How to Compare LDA Models¶
Demonstrates how you can visualize and compare trained topic models.
# sphinx_gallery_thumbnail_number = 2
import logging
logging.basicConfig(format='%(asctime)s : %(levelname)s : %(message)s', level=logging.INFO)
First, clean up the 20 Newsgroups dataset. We will use it to fit LDA.¶
from string import punctuation
from nltk import RegexpTokenizer
from nltk.stem.porter import PorterStemmer
from nltk.corpus import stopwords
from sklearn.datasets import fetch_20newsgroups
newsgroups = fetch_20newsgroups()
eng_stopwords = set(stopwords.words('english'))
tokenizer = RegexpTokenizer(r'\s+', gaps=True)
stemmer = PorterStemmer()
translate_tab = {ord(p): u" " for p in punctuation}
def text2tokens(raw_text):
"""Split the raw_text string into a list of stemmed tokens."""
clean_text = raw_text.lower().translate(translate_tab)
tokens = [token.strip() for token in tokenizer.tokenize(clean_text)]
tokens = [token for token in tokens if token not in eng_stopwords]
stemmed_tokens = [stemmer.stem(token) for token in tokens]
return [token for token in stemmed_tokens if len(token) > 2] # skip short tokens
dataset = [text2tokens(txt) for txt in newsgroups['data']] # convert a documents to list of tokens
from gensim.corpora import Dictionary
dictionary = Dictionary(documents=dataset, prune_at=None)
dictionary.filter_extremes(no_below=5, no_above=0.3, keep_n=None) # use Dictionary to remove un-relevant tokens
dictionary.compactify()
d2b_dataset = [dictionary.doc2bow(doc) for doc in dataset] # convert list of tokens to bag of word representation
Second, fit two LDA models.¶
from gensim.models import LdaMulticore
num_topics = 15
lda_fst = LdaMulticore(
corpus=d2b_dataset, num_topics=num_topics, id2word=dictionary,
workers=4, eval_every=None, passes=10, batch=True,
)
lda_snd = LdaMulticore(
corpus=d2b_dataset, num_topics=num_topics, id2word=dictionary,
workers=4, eval_every=None, passes=20, batch=True,
)
Time to visualize, yay!¶
We use two slightly different visualization methods depending on how you’re running this tutorial. If you’re running via a Jupyter notebook, then you’ll get a nice interactive Plotly heatmap. If you’re viewing the static version of the page, you’ll get a similar matplotlib heatmap, but it won’t be interactive.
def plot_difference_plotly(mdiff, title="", annotation=None):
"""Plot the difference between models.
Uses plotly as the backend."""
import plotly.graph_objs as go
import plotly.offline as py
annotation_html = None
if annotation is not None:
annotation_html = [
[
"+++ {}<br>--- {}".format(", ".join(int_tokens), ", ".join(diff_tokens))
for (int_tokens, diff_tokens) in row
]
for row in annotation
]
data = go.Heatmap(z=mdiff, colorscale='RdBu', text=annotation_html)
layout = go.Layout(width=950, height=950, title=title, xaxis=dict(title="topic"), yaxis=dict(title="topic"))
py.iplot(dict(data=[data], layout=layout))
def plot_difference_matplotlib(mdiff, title="", annotation=None):
"""Helper function to plot difference between models.
Uses matplotlib as the backend."""
import matplotlib.pyplot as plt
fig, ax = plt.subplots(figsize=(18, 14))
data = ax.imshow(mdiff, cmap='RdBu_r', origin='lower')
plt.title(title)
plt.colorbar(data)
try:
get_ipython()
import plotly.offline as py
except Exception:
#
# Fall back to matplotlib if we're not in a notebook, or if plotly is
# unavailable for whatever reason.
#
plot_difference = plot_difference_matplotlib
else:
py.init_notebook_mode()
plot_difference = plot_difference_plotly
Gensim can help you visualise the differences between topics. For this purpose, you can use the diff() method of LdaModel.
diff() returns a matrix with distances mdiff and a matrix with annotations annotation. Read the docstring for more detailed info.
In each mdiff[i][j] cell you’ll find a distance between topic_i from the first model and topic_j from the second model.
In each annotation[i][j] cell you’ll find [tokens from intersection, tokens from difference between topic_i from first model and topic_j from the second model.
print(LdaMulticore.diff.__doc__)
Out:
Calculate the difference in topic distributions between two models: `self` and `other`.
Parameters
----------
other : :class:`~gensim.models.ldamodel.LdaModel`
The model which will be compared against the current object.
distance : {'kullback_leibler', 'hellinger', 'jaccard', 'jensen_shannon'}
The distance metric to calculate the difference with.
num_words : int, optional
The number of most relevant words used if `distance == 'jaccard'`. Also used for annotating topics.
n_ann_terms : int, optional
Max number of words in intersection/symmetric difference between topics. Used for annotation.
diagonal : bool, optional
Whether we need the difference between identical topics (the diagonal of the difference matrix).
annotation : bool, optional
Whether the intersection or difference of words between two topics should be returned.
normed : bool, optional
Whether the matrix should be normalized or not.
Returns
-------
numpy.ndarray
A difference matrix. Each element corresponds to the difference between the two topics,
shape (`self.num_topics`, `other.num_topics`)
numpy.ndarray, optional
Annotation matrix where for each pair we include the word from the intersection of the two topics,
and the word from the symmetric difference of the two topics. Only included if `annotation == True`.
Shape (`self.num_topics`, `other_model.num_topics`, 2).
Examples
--------
Get the differences between each pair of topics inferred by two models
.. sourcecode:: pycon
>>> from gensim.models.ldamulticore import LdaMulticore
>>> from gensim.test.utils import datapath
>>>
>>> m1 = LdaMulticore.load(datapath("lda_3_0_1_model"))
>>> m2 = LdaMulticore.load(datapath("ldamodel_python_3_5"))
>>> mdiff, annotation = m1.diff(m2)
>>> topic_diff = mdiff # get matrix with difference for each topic pair from `m1` and `m2`
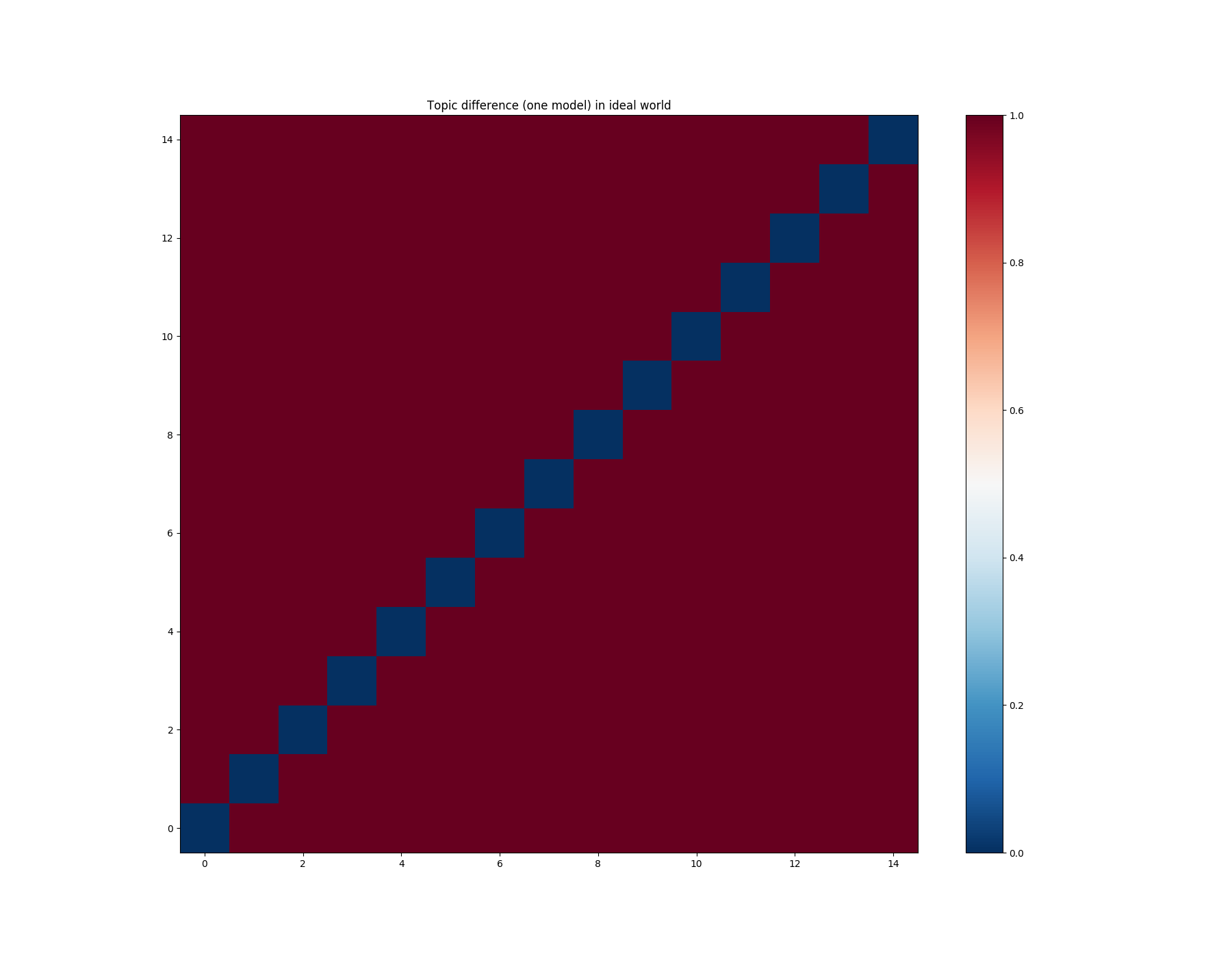
Case 1: How topics within ONE model correlate with each other.¶
Short description:
x-axis - topic;
y-axis - topic;
almost red cell - strongly decorrelated topics;
almost blue cell - strongly correlated topics.
In an ideal world, we would like to see different topics decorrelated between themselves. In this case, our matrix would look like this:
import numpy as np
mdiff = np.ones((num_topics, num_topics))
np.fill_diagonal(mdiff, 0.)
plot_difference(mdiff, title="Topic difference (one model) in ideal world")
Unfortunately, in real life, not everything is so good, and the matrix looks different.
Short description (interactive annotations only):
+++ make, world, well- words from the intersection of topics = present in both topics;--- money, day, still- words from the symmetric difference of topics = present in one topic but not the other.
mdiff, annotation = lda_fst.diff(lda_fst, distance='jaccard', num_words=50)
plot_difference(mdiff, title="Topic difference (one model) [jaccard distance]", annotation=annotation)
![Topic difference (one model) [jaccard distance]](../../_images/sphx_glr_run_compare_lda_002.png)
If you compare a model with itself, you want to see as many red elements as possible (except on the diagonal). With this picture, you can look at the “not very red elements” and understand which topics in the model are very similar and why (you can read annotation if you move your pointer to cell).
Jaccard is a stable and robust distance function, but sometimes not sensitive enough. Let’s try to use the Hellinger distance instead.
mdiff, annotation = lda_fst.diff(lda_fst, distance='hellinger', num_words=50)
plot_difference(mdiff, title="Topic difference (one model)[hellinger distance]", annotation=annotation)
![Topic difference (one model)[hellinger distance]](../../_images/sphx_glr_run_compare_lda_003.png)
You see that everything has become worse, but remember that everything depends on the task.
Choose a distance function that matches your upstream task better: what kind of “similarity” is relevant to you. From my (Ivan’s) experience, Jaccard is fine.
Case 2: How topics from DIFFERENT models correlate with each other.¶
Sometimes, we want to look at the patterns between two different models and compare them.
You can do this by constructing a matrix with the difference.
mdiff, annotation = lda_fst.diff(lda_snd, distance='jaccard', num_words=50)
plot_difference(mdiff, title="Topic difference (two models)[jaccard distance]", annotation=annotation)
![Topic difference (two models)[jaccard distance]](../../_images/sphx_glr_run_compare_lda_004.png)
Looking at this matrix, you can find similar and different topics between the two models. The plot also includes relevant tokens describing the topics’ intersection and difference.
Total running time of the script: ( 3 minutes 55.324 seconds)
Estimated memory usage: 303 MB
